Demo¶
[1]:
import numpy as np
import scipy as sp
import scipy.sparse
import matplotlib.pyplot as plt
%matplotlib inline
[2]:
from bayesbridge import BayesBridge, RegressionModel, RegressionCoefPrior
from simulate_data import simulate_design, simulate_outcome
from util import mcmc_summarizer
BayesBridge supports both dense (numpy array) and sparse (scipy sparse matrix) design matrices.¶
[3]:
n_obs, n_pred = 10 ** 4, 10 ** 3
X = simulate_design(
n_obs, n_pred,
binary_frac=.9,
binary_pred_freq=.2,
shuffle_columns=True,
format_='sparse',
seed=111
)
[4]:
beta_true = np.zeros(n_pred)
beta_true[:5] = 1.5
beta_true[5:10] = 1.
beta_true[10:15] = .5
n_trial = np.ones(X.shape[0]) # Binary outcome.
y = simulate_outcome(
X, beta_true, intercept=0.,
n_trial=n_trial, model='logit', seed=1
)
Specify regression model and prior.¶
BayesBridge uses a prior \(\pi(\beta_j \, | \, \tau) \propto \tau^{-1} \exp\left( - \, \left| \tau^{-1} \beta_j \right|^\alpha \right)\) for \(0 < \alpha \leq 1\). The default bridge exponent is \(\alpha = 1 / 2\).
[5]:
model = RegressionModel(
y, X, family='logit',
add_intercept=True, center_predictor=True,
# Do *not* manually add intercept to or center X.
)
prior = RegressionCoefPrior(
bridge_exponent=.5,
n_fixed_effect=0,
# Number of coefficients with Gaussian priors of pre-specified sd.
sd_for_intercept=float('inf'),
# Set it to float('inf') for a flat prior.
sd_for_fixed_effect=1.,
regularizing_slab_size=2.,
# Weakly constrain the magnitude of coefficients under bridge prior.
)
bridge = BayesBridge(model, prior)
Run the Gibbs sampler.¶
[6]:
samples, mcmc_info = bridge.gibbs(
n_iter=250, n_burnin=0, thin=1,
init={'global_scale': .01},
coef_sampler_type='cg',
seed=111
)
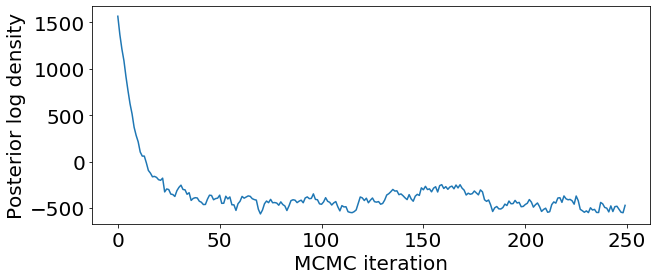
Check convergence by looking at the traceplot for posterior log-density.
[7]:
plt.figure(figsize=(10, 4))
plt.rcParams['font.size'] = 20
plt.plot(samples['logp'])
plt.xlabel('MCMC iteration')
plt.ylabel('Posterior log density')
plt.show()
Restart MCMC from the last iteration with ‘gibbs_additional_iter()’.¶
[8]:
samples, mcmc_info = bridge.gibbs_additional_iter(
mcmc_info, n_add_iter=250
)
Add more samples (while keeping the previous ones) with ‘merge=True’.
[9]:
samples, mcmc_info = bridge.gibbs_additional_iter(
mcmc_info, n_add_iter=750, merge=True, prev_samples=samples
)
coef_samples = samples['coef'][1:, :] # Extract all but the intercept
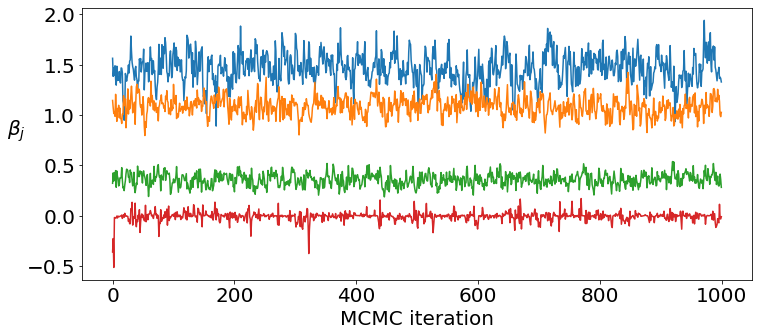
Check mixing of regression coefficients and their posterior marginals.¶
Typically the convergence is quick and mixing of the regression coefficients is adequate.
[10]:
plt.figure(figsize=(12, 5))
plt.rcParams['font.size'] = 20
plt.plot(coef_samples[[0, 5, 10, 15], :].T)
plt.xlabel('MCMC iteration')
plt.ylabel(r'$\beta_j$', rotation=0, labelpad=10)
plt.show()
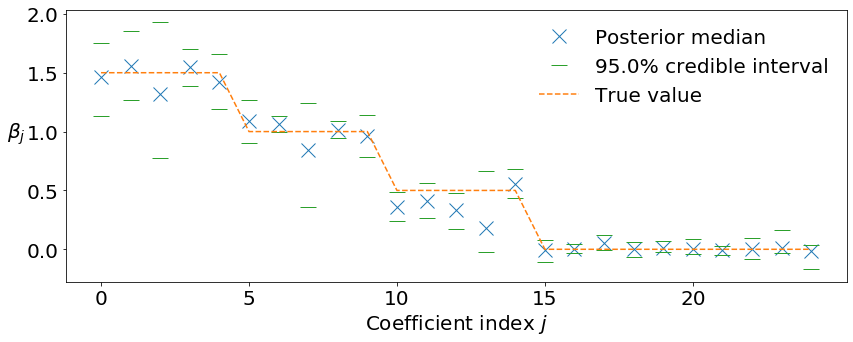
[11]:
plt.figure(figsize=(14, 5))
plt.rcParams['font.size'] = 20
n_coef_to_plot = 25
mcmc_summarizer.plot_conf_interval(
coef_samples, conf_level=.95,
n_coef_to_plot=n_coef_to_plot, marker_scale=1.4
);
plt.plot(
beta_true[:n_coef_to_plot], '--', color='tab:orange',
label='True value'
)
plt.xlabel(r'Coefficient index $j$')
plt.ylabel(r'$\beta_j$', rotation=0, labelpad=10)
plt.xticks([0, 5, 10, 15, 20])
plt.legend(frameon=False)
plt.show()